 |
About complex I
The mitochondrial complex I (NADH-quinone oxidoreductase, EC 1.6.5.3 or proton-pumping NADH dehydrogenase Type 1) is one of three energy-transducing enzyme complexes of the respiratory chain in mitochondria. This enzyme catalyses the reaction:
NADH + Q + H+ + 4H+in -> NAD+ + QH2 + 4H+out (Q=ubiquinone)
Complex I is the point of entry for the major fraction of electrons that traverse the respiratory chain eventually resulting in the reduction of oxygen. Coupled to the electron transfer, protons are pumped from the matrix side to the intermembrane space of mitochondria. It is the most intricate membrane-bound enzyme known to date, being composed of at least 45 different polypeptides. Of these, 7 are encoded by mitochondrial DNA.
The first isolation of complex I from bovine heart mitochondria by deoxycholate fractionation (PMID:13905327) by Joe Hatefi and coworkers has been conducted at the beginning of 60's, but until recently, information on its structure and mechanism of action was still limited. This enzyme complex contains a noncovalently-bound FMN molecule and 2 binuclear and 6 tetranuclear iron-sulfur (FeS) clusters, of which 2 binuclear and 4 tetranuclear clusters are EPR-visible.
Why research on complex I is important
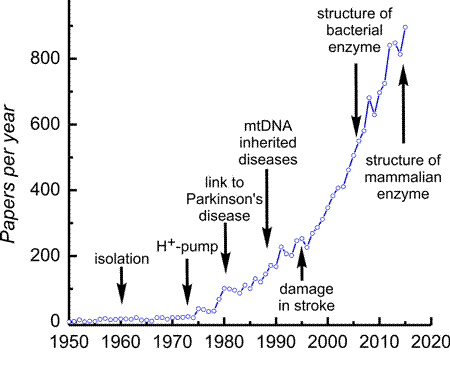 Research on complex I has recently taken on greater significance since the finding that many human inherited mitochondrial diseases involve structural and functional defects at the level of this enzyme. Many cases of Leber's hereditary optic neuropathy (LHON), Mitochondrial Encephalomyopathy Lactic Acidosis, and Stroke-like episodes (MELAS) and Leigh syndrome appear to be associated with defects in complex I (PMID:27775730). In addition to these documented cases, complex I inhibitors, 1-methyl-4-phenylpyridium and rotenone, produce drug-induced Parkinsonism in rodents and human, which suggests a link between Parkinson's disease and function of mitochondrial complex I (PMID:22229791). In many models of cardiovascular pathologies associated with ischemia/reperfusion (cardiac infarction and stroke) complex I was shown to be the most vulnerable enzyme that is damaged first. Complex I is also known to be a target of new pesticides such as acaricides. Moreover, it is suggested that complex I malfunction is involved in the pathogenesis of diabetes. Therefore, complex I research is important to not only basic sciences but also clinical medicine. This is evident in the progressively growing number of publications relating to complex I, as depicted in the figure. There are several milestones in complex I research. In 1973, Ragan and Racker showed that complex I is a proton pump. In the mid-1980s, it was found that the 7 unidentified genes in mtDNA are encoding complex I subunits by Attardi's group. Shortly after that, the first case of mitochondrial diseases caused by complex I defects was reported (Wallace et al., 1988). Following the first publication from Sazanov's lab on the architecture of the hydrophilic domain of bacterial enzyme (PMID: 16469879) in 2006, the structure of mammalian enzyme has been resolved almost completely and published by Hirst's and Sazanov's groups in 2016 (PMID:27595392; PMID: 27509854, also see Research section). Research on complex I has recently taken on greater significance since the finding that many human inherited mitochondrial diseases involve structural and functional defects at the level of this enzyme. Many cases of Leber's hereditary optic neuropathy (LHON), Mitochondrial Encephalomyopathy Lactic Acidosis, and Stroke-like episodes (MELAS) and Leigh syndrome appear to be associated with defects in complex I (PMID:27775730). In addition to these documented cases, complex I inhibitors, 1-methyl-4-phenylpyridium and rotenone, produce drug-induced Parkinsonism in rodents and human, which suggests a link between Parkinson's disease and function of mitochondrial complex I (PMID:22229791). In many models of cardiovascular pathologies associated with ischemia/reperfusion (cardiac infarction and stroke) complex I was shown to be the most vulnerable enzyme that is damaged first. Complex I is also known to be a target of new pesticides such as acaricides. Moreover, it is suggested that complex I malfunction is involved in the pathogenesis of diabetes. Therefore, complex I research is important to not only basic sciences but also clinical medicine. This is evident in the progressively growing number of publications relating to complex I, as depicted in the figure. There are several milestones in complex I research. In 1973, Ragan and Racker showed that complex I is a proton pump. In the mid-1980s, it was found that the 7 unidentified genes in mtDNA are encoding complex I subunits by Attardi's group. Shortly after that, the first case of mitochondrial diseases caused by complex I defects was reported (Wallace et al., 1988). Following the first publication from Sazanov's lab on the architecture of the hydrophilic domain of bacterial enzyme (PMID: 16469879) in 2006, the structure of mammalian enzyme has been resolved almost completely and published by Hirst's and Sazanov's groups in 2016 (PMID:27595392; PMID: 27509854, also see Research section).
Mammalian complex I
Mammalian complex I is composed of 45 different subunits and contains noncovalently bound FMN and several iron-sulfur clusters as prosthetic groups. On the contrary, S. cerevisiae does not have complex I but has membrane-associated rotenone-insensitive NADH-quinone oxidoreductases (Ndi1) which are composed of a single subunit and FAD as cofactor and no iron-sulfur clusters. In mitochondria of certain fungi and plants, complex I and rotenone-insensitive NADH-quinone oxidoreductases coexist.
Bacterial NADH dehydrogenases
The NADH-quinone oxidoreductases of the bacterial respiratory chains can be divided into at least three groups (see TABLE). Those are the proton-translocating NADH-quinone oxidoreductase (NDH-1), the NADH-quinone oxidoreductase lacking an energy coupling site (NDH-2, very close to Ndi1), and Na+-translocating NADH-quinone oxidoreductase (Na+-NDH). The NDH-1 is composed of 14 different subunits, suggesting that the NDH-1 has relatively simpler structure than complex I. It pumps protons from the cytosolic side to the periplasmic side. The NDH-2 is a single subunit enzyme and bears non-covalently bound FAD and has no iron-sulfur clusters similar to mitochondrial rotenone-insensitive NADH-quinone oxidoreductases. Na+-NDH is made up of 6 unlike subunits and contains two covalently bound FMN, one noncovalently bound FAD, one noncovalently bound riboflavin, and one binuclear iron-sulfur cluster. In terms of cofactors and specific inhibitors, NDH-1 is akin to complex I.
|
 |

